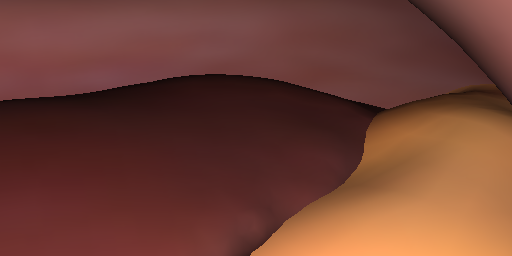
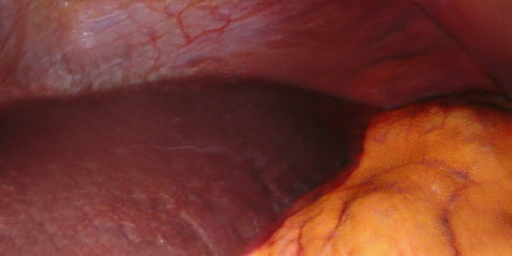


We designed a method for rendering long-term temporally consistent and realistic video sequences from simulated surgical 3D scenes. By generating realistic data from simulations, we are able to provide rich ground truth which would otherwise be very challenging to obtain (e.g. camera poses, 3D geometry, point correspondences). This way, we provide data that is potentially useful for evaluating or training tasks such as SLAM, visual servoing or tracking in a surgical setting. On this page, video examples, code and our complete generated dataset can be found.
| Download | Details | Comments | Size | |
|---|---|---|---|---|
 |
Reference (Input) Images | aref, Simple renderings of 21 000 random views (7 simulated scenes with 3000 views each). | These reference images were used as partial input to the translation module (next to the projected texture features). | ca. 1.3 GB |
 |
Translations | b̂, Realistic, view-consistent translation of each view. | The model was trained to render textures with consistent hue while allowing for varying brightness (view-dependent effects). | ca. 3.2 GB |
 |
Labels: Segmentation | Full segmentation masks. | Classes: liver (blue), fat/stomach (red), abdominal wall (green), gallbladder (cyan), ligament (magenta). | ca. 25 MB |
 |
Labels: Depth | Depth maps. | Stored as HDR gray-scale images. | ca. 1 GB |
 |
Labels: 3D-Coordinates | Corresponding 3D surface coordinates for each pixel. | Stored as HDR color images. Can be used to obtain dense pixel correspondences between views (e.g. for optical flow, tracking, SLAM). The model was trained to render textures with consistent hue while allowing for varying brightness (view-dependent effects). See the demo for usage. | ca. 25 GB |
 |
Labels: Camera Poses | Camera poses. | Stored as 7D vectors (3 for location, 4 for quaterion rotation). See the demo for usage. | ca. 800 kB |
| Download | Details | Comments | Size | |
|---|---|---|---|---|
 |
Reference Images (Sequences) | aref, Simple renderings of 11 sequences (100 frames at 5fps each). | These reference images were used as partial input to the translation module (next to the projected texture features). | ca. 60 MB |
 |
Translations (Sequences) | b̂, Realistic, view-consistent translation of each view. | The model was trained to render textures with consistent hue while allowing for varying brightness (view-dependent effects). | ca. 160 MB |
 |
Labels: Segmentation (Sequences) | Full segmentation masks. | Classes: liver (blue), fat/stomach (red), abdominal wall (green), gallbladder (cyan), ligament (magenta). | ca. 1.5 MB |
 |
Labels: Depth (Sequences) | Depth maps. | Stored as HDR gray-scale images. | ca. 50 MB |
 |
Labels: 3D-Coordinates (Sequences) | Corresponding 3D surface coordinates for each pixel. | Stored as HDR color images. Can be used to obtain dense pixel correspondences between views (e.g. for optical flow, tracking, SLAM). The model was trained to render textures with consistent hue while allowing for varying brightness (view-dependent effects). See the demo for usage. | ca. 1.3 GB |
 |
Labels: Camera Poses | Camera poses. | Stored as 7D vectors (3 for location, 4 for quaterion rotation). See the demo for usage. | ca. 35 kB |
| Simulated Scene 6 | Simulated Scene 5 |
|---|---|
| Simulated Scene 1 | Simulated Scene 1 |
| Simulated Scene 3 | |
If you use our data or code, please cite the following paper:
"Long-Term Temporally Consistent Unpaired Video Translation from Simulated Surgical 3D Data".The code for this project can be found on GitLab.
It is based on Pfeiffer et al.'s framework for surgical image translation (Code, Paper) and the Multimodal UNsupervised Image-to-image Translation (MUNIT) framework (Code, Paper).
| NCT/UCC Dresden | Centre for Tactile Internet, TU Dresden | University Hospital Dresden | German Cancer Research Center |