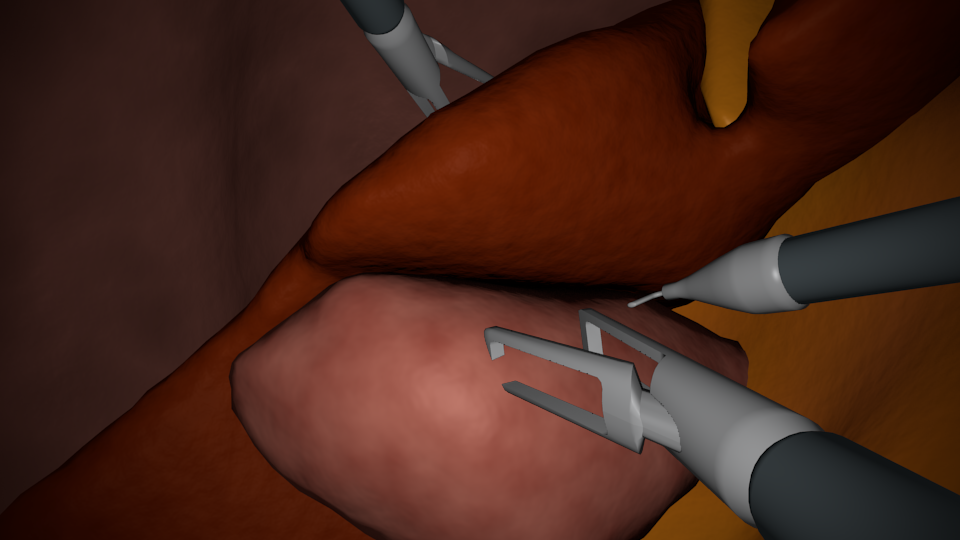
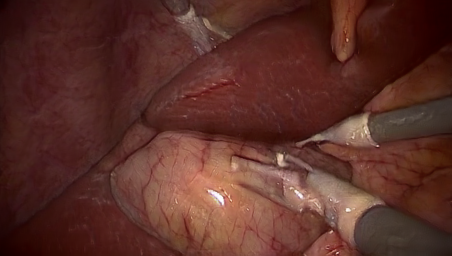
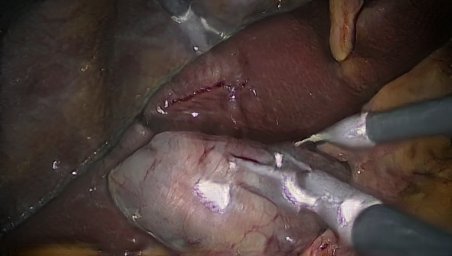

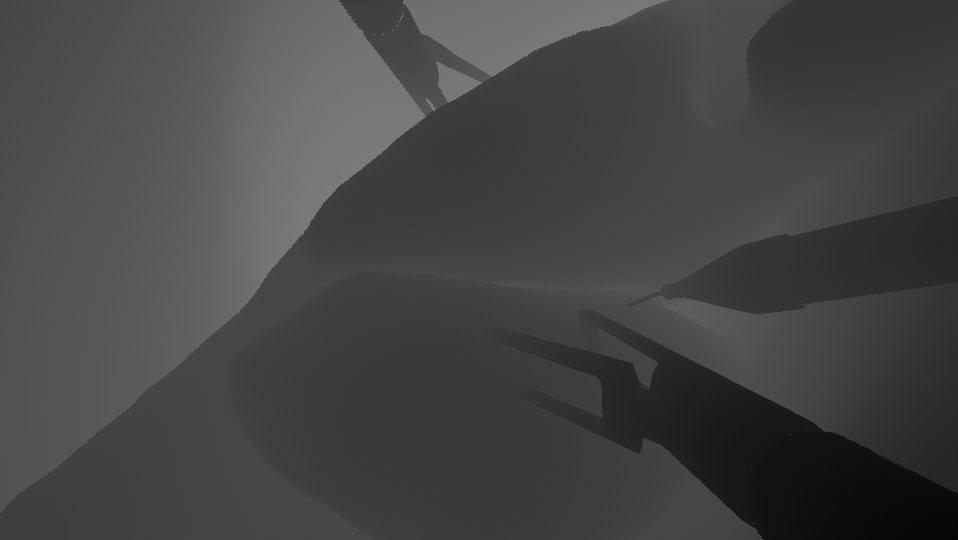


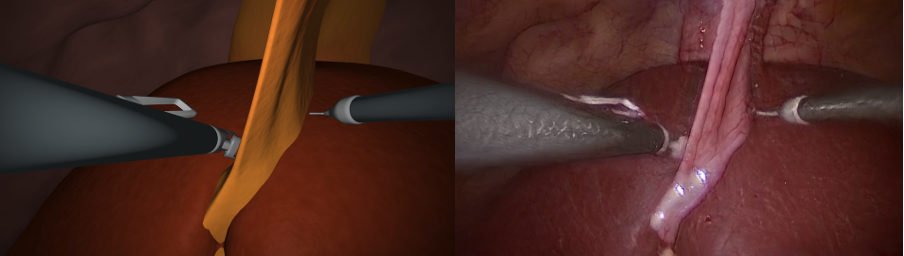
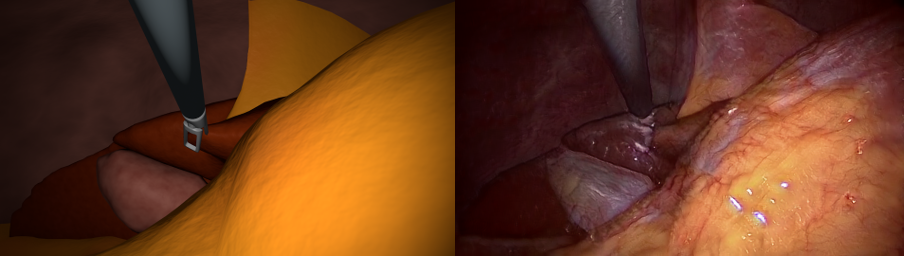
In this project, we generate synthetic images in a 3D environemnt, roughly resembling laparoscopic liver surgery scenes. We then train a group of Generative Adversarial Networks (GAN) to translate these images to look like real laparoscopic images. After the training process, we can use the translated images along with their labels as training data for a certain target task. The data sets as well as the code to generate them are made publically available (see below).
If you use our data or code, please cite the following paper:
"Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation". Micha Pfeiffer, Isabel Funke, Maria R. Robu, Sebastian Bodenstedt, Leon Strenger, Sandy Engelhardt, Tobias Roß, Matthew J. Clarkson, Kurinchi Gurusamy, Brian R. Davidson, Lena Maier-Hein, Carina Riediger, Thilo Welsch, Jürgen Weitz, and Stefanie Speidel. MICCAI 2019.| Name | Details | Comments | |
|---|---|---|---|
 |
Input Images | A+, 20 000 images | These are the input images in the simulated domain which we gave to the translator. Usually not required, unless you want to train your own translator. |
 |
Translations, Random stlye | Bsyn, Random style vectors, 100 000 Images | Translations of the Input Images. Each input image was translated five times, each time with a new randomly sampled style. These kind of images were used to obtain the results in our paper. |
 |
Translations, Cholec80 style | Alternative translation. Style vectors extracted from Cholec80, 100 000 Images | Translations of the Input Images. Each input image was translated five times, each time with a new style extracted from a random Cholec80 image b. At this point, we cannot say whether this data set or the random style data set is better for training networks. You may want to try both, or even mix the two to increase diversity in style. |
 |
Labels: Segmentation | Full segmentation masks | Segmentation mask for every image in A+. Classes are: Liver, Fat, Abdominal Wall, Tool Shaft, Tool Tip, Gallbladder |
 |
Labels: Depth | Depth maps | Depth map for every image in A+. |
 |
Labels: Ridges | Ridge Lines | Liver ridge lines for every image. The ridges were extracted semi-automatically from the 3D liver meshes. Afterwards, they were rendered into the camera view to obtain these images. As the ridges follow surface edges, they may not be smooth everywhere. Please use post-processing like thickening and smoothing where necessary. |
 |
Labels: Normals | Normals | Surface normals in world space. Please use the camera information to transform these to camera space if needed. |
| Labels: Camera Information (Coming soon) | Coming soon | Coming soon |
The code for this project can be found on GitLab.
It is based on the Multimodal UNsupervised Image-to-image Translation (MUNIT) framework: Code, Paper
| NCT Dresden | University Clinic Dresden | German Cancer Research Center |